6.1 Simple Temporal Effects
We might have measured a variable over the course of a certain time period (e.g. 20 years). We might expect that there is stochastic year-to-year variation, which we can simulate already. However we might also want to simulate patterns in that temporal data. We can treat the levels associated with a particular grouping factor (e.g. year) as both a factor and continuous.
To treat a grouping factor as continuous, we use covariate=TRUE in the parameter list. In this way we can simulate a linear effect of year:
squid_data <- simulate_population(
data_structure= make_structure(structure = "year(20) + sex(2)/individual(50)",repeat_obs=20),
parameters=list(
year_cont = list(
group="year",
names= "year_cont",
covariate=TRUE,
beta=0.3
),
year = list(
vcov = 0.8
),
residual=list(
vcov = 1
)
)
)note we have specified group in the parameter list. This enables us to link a set of parameters to the grouping factor in the data structure. This doesn’t have to be specified and defaults to the name of the list item.
data <- get_population_data(squid_data)
head(data)## y year_cont year_effect residual year sex individual squid_pop
## 1 1.1436403 1 2.21821 -1.3745698 1 1 1 1
## 2 3.0559676 1 2.21821 0.5377575 1 1 1 1
## 3 4.2148492 1 2.21821 1.6966392 1 1 1 1
## 4 0.6152528 1 2.21821 -1.9029573 1 1 1 1
## 5 1.8172122 1 2.21821 -0.7009978 1 1 1 1
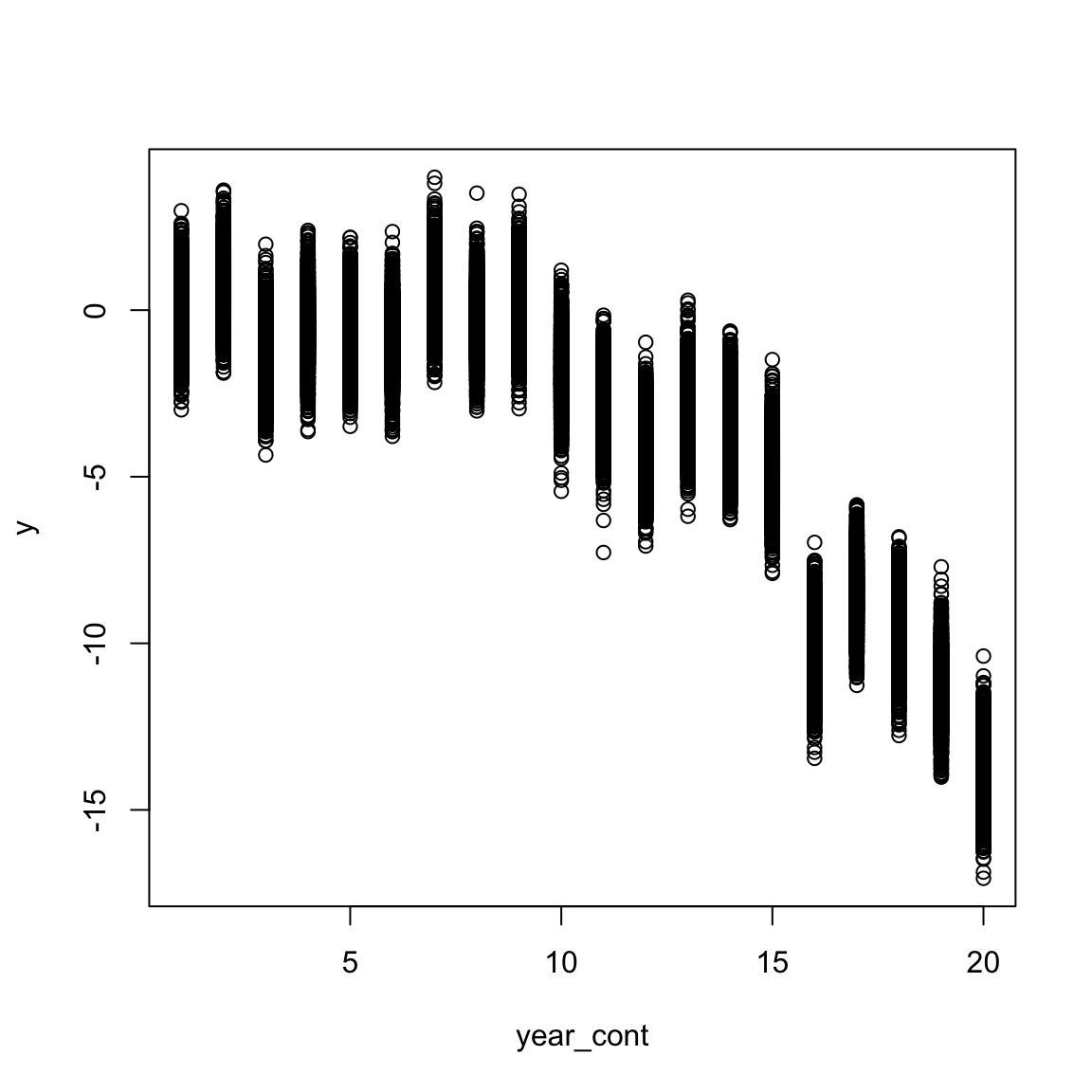
## 6 1.7139274 1 2.21821 -0.8042827 1 1 1 1plot(y ~ year_cont, data)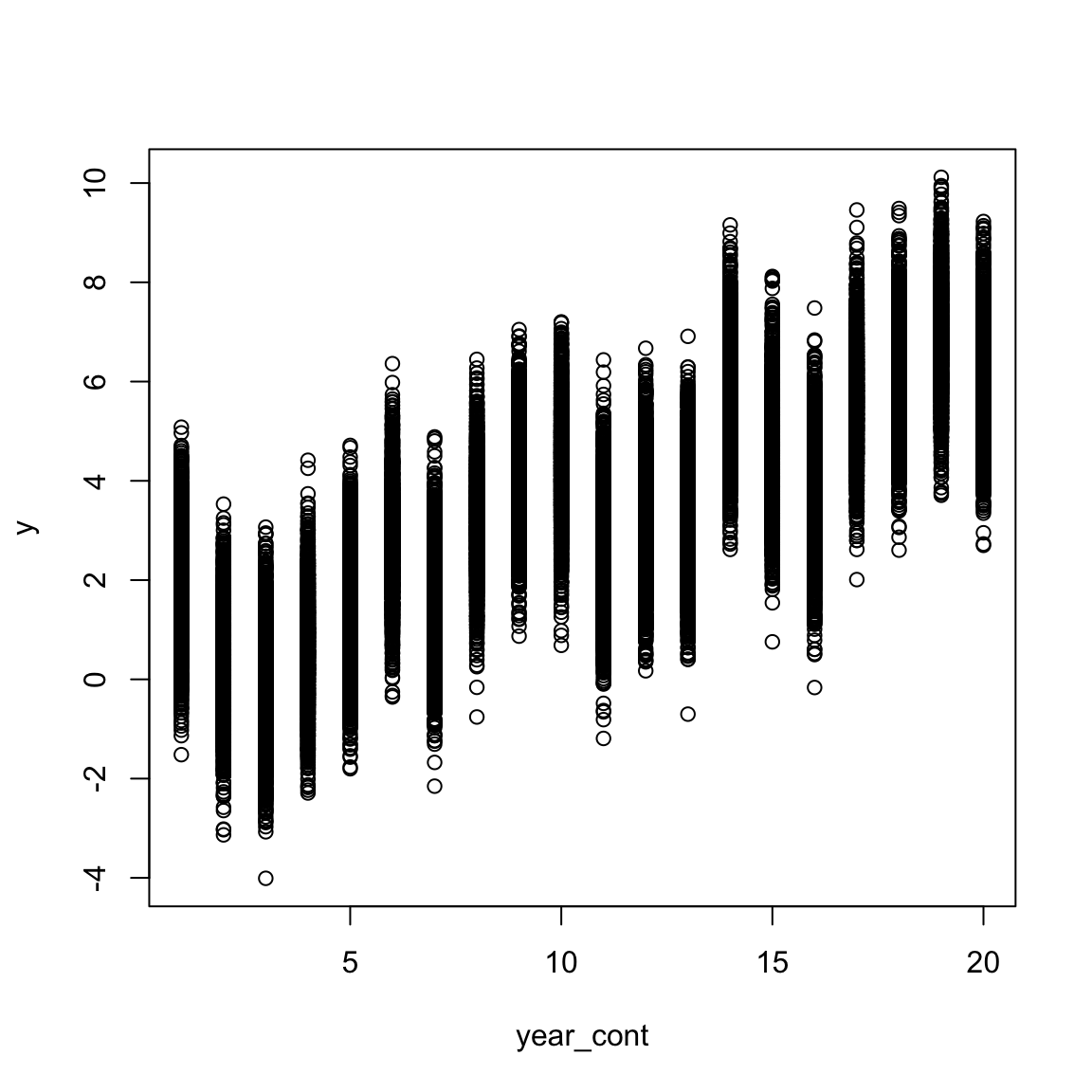
Here we can see there is within year variation, year to year variation, as well as a linear directional year effect.
lmer(y ~ year_cont + (1|year), data)## Linear mixed model fit by REML ['lmerMod']
## Formula: y ~ year_cont + (1 | year)
## Data: data
## REML criterion at convergence: 113318.3
## Random effects:
## Groups Name Std.Dev.
## year (Intercept) 0.9652
## Residual 0.9956
## Number of obs: 40000, groups: year, 20
## Fixed Effects:
## (Intercept) year_cont
## 0.5669 0.2838In a similar way we can also simulate a quadratic effect of time.
squid_data <- simulate_population(
data_structure = make_structure(structure = "year(20) + sex(2)/individual(50)",repeat_obs=20),
parameters=list(
year_cont = list(
group="year",
names= c("year_cont"),
covariate=TRUE,
beta=c(0.3)
),
interactions=list(
names= c("year_cont:year_cont"),
beta=c(-0.05)
),
year = list(
vcov = 1
),
residual=list(
vcov = 0.8
)
)
)
data <- get_population_data(squid_data)
plot(y~year_cont,data)