1.3 Interactions and non-linear effects
1.3.1 Interactions
\[ y_i = \beta_0 + \beta_1 x_{1i} + \beta_2 x_{2i} + \beta_3 x_{1i}x_{2i} + \epsilon_i \]
We can specify the interaction between two predictors by adding an interactions list to the parameters list. Interactions can then be specified between two named variables using “:”. Interactions can be specified between two predictors at the same level, or at different hierarchical levels.
squid_data <- simulate_population(
n=2000,
response_name = "body_mass",
parameters=list(
observation=list(
names=c("temperature","rainfall"),
beta = c(0.5,0.3)
),
residual=list(
vcov=0.3
),
interactions=list(
names=c("temperature:rainfall"),
beta = c(-0.1)
)
)
)
data <- get_population_data(squid_data)
head(data)## body_mass temperature rainfall residual temperature:rainfall squid_pop
## 1 -0.1950863 0.6545190 1.5577734 -0.8877186 1.0195922 1
## 2 -0.8352969 -1.3405423 0.3989025 -0.3381711 -0.5347457 1
## 3 -0.2615434 -1.2677241 -0.2484353 0.4783440 0.3149474 1
## 4 0.8427175 0.8669511 0.1360867 0.3802140 0.1179805 1
## 5 -0.8451921 -1.8557505 0.8777406 -0.3435258 -1.6288676 1
## 6 0.7969244 -0.2192104 0.8250010 0.6409444 -0.1808488 1coef(lm(body_mass ~ temperature * rainfall, data))## (Intercept) temperature rainfall
## 0.006055206 0.487910683 0.284464667
## temperature:rainfall
## -0.0843422681.3.2 Non-linear effects
Polynomial (quadratic, cubic, etc) functions are essentially interactions with the same predictor. They can therefore be specified in the same way:
squid_data <- simulate_population(
n=2000,
response_name = "body_mass",
parameters=list(
observation=list(
names=c("temperature"),
beta = c(0.5)
),
interactions=list(
names=c("temperature:temperature"),
beta = c(-0.3)
),
residual=list(
vcov=0.3
)
)
)
data <- get_population_data(squid_data)
plot(body_mass ~ temperature, data, pch=19, cex=0.5, col=alpha(1,0.5))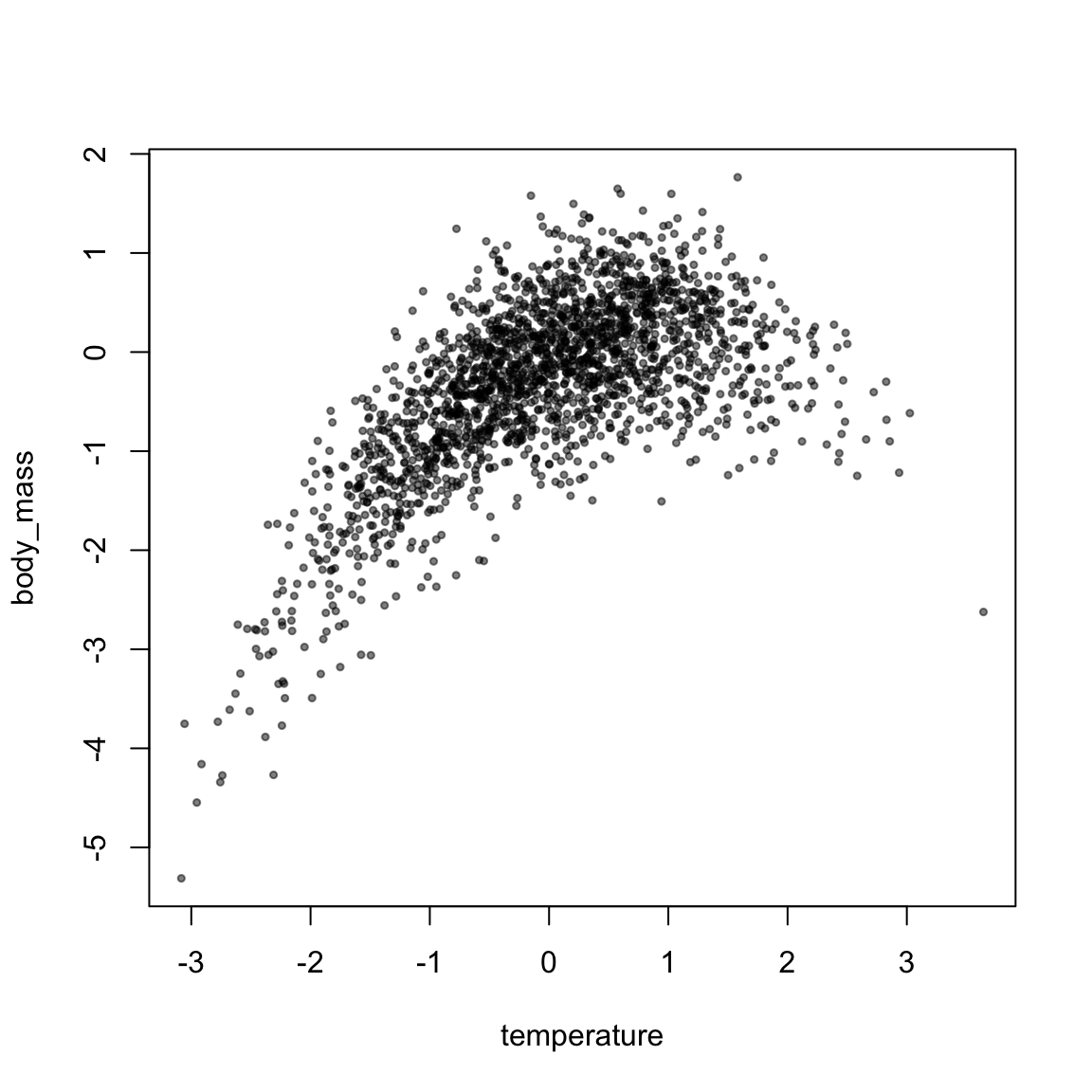
coef(lm(body_mass ~ temperature + I(temperature^2), data))## (Intercept) temperature I(temperature^2)
## -0.003753765 0.510987516 -0.299719778